Ordering
*link will take you to our exclusive distribution partner site
*link will take you to our exclusive distribution partner site
MyTaq™ HS Red Mix is recommended for PCR assays containing complex and low copy number targets as well as multiplex PCR. MyTaq HS Red Mix is comprised of MyTaq HS DNA Polymerase and a novel buffer system that deliver very high yield PCR amplification over a wide range of PCR templates. MyTaq HS has an increased affinity for DNA, enabling reliable amplification from even very low amounts of template. MyTaq HS has been developed to give more robust amplification than other commonly-used polymerases allowing it to perform well with challenging templates and in the presence of PCR inhibitors. Furthermore, the highly efficient nature of MyTaq HS means it gives excellent results under fast PCR conditions. MyTaq HS does not possess polymerase activity during the reaction set-up, thereby reducing the non-specific amplification that can hinder PCR assays from the start.
The product is supplied as a mastermix that requires the addition of only template, primers and water, thereby reducing the number of pipetting steps during PCR set-up for improved speed, throughput and assay reproducibility. The inclusion of dNTPs, MgCl2 and enhancers at optimal concentrations, helps eliminate the need for optimization, thereby saving time and ensuring comparable efficiencies for annealing and extension of all primers in the reaction, this makes MyTaq HS Red Mix ideal for multiplex PCR.
MyTaq HS Red Mix, includes a red dye that increases the visual contrast between the reagent and the reaction vessel for improved convenience and to improve pipetting accuracy. The red dye also enables samples to be loaded directly on to a gel after the PCR eliminating the need to add loading buffer.
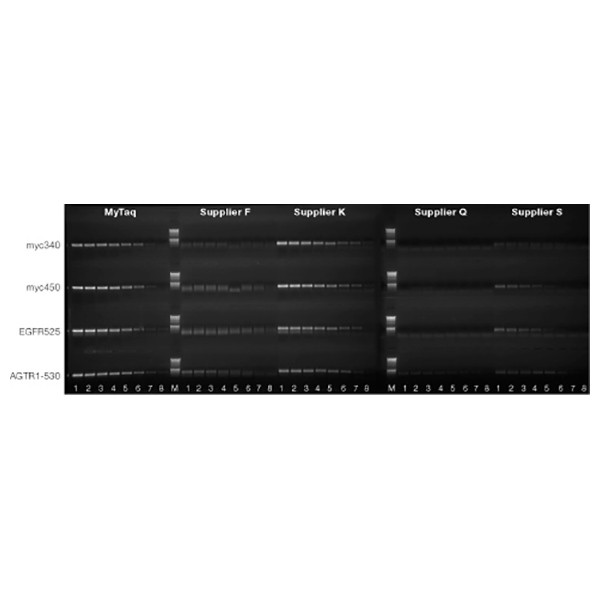
A 340 bp (A) and a 450 bp (B) fragment of the myc gene, a 525 bp (C) fragment of the EGFR gene and a 530 bp (D) fragment of the AGRI1 gene were amplified using MyTaq HS and hot-start DNA polymerases from Suppliers F, K, G and S. Each polymerase was used to set-up PCR reactions containing either 100 ng, 33 ng, 10 ng, 4 ng, 1 ng, 33 pg, 10 pg and 3 pg of human genomic DNA (Lanes 1-8 respectively), prepared by a 3-fold serial dilution. Marker is HyperLadder 1kb (M). MyTaq HS performed well across all four human genes.
M13 vector carrying a DNA insert was cloned into E.coli cells. 2 µL increments of agar (2a) and 2 µL increments of LB (2b) were added to a series of 50 µL PCR reactions (Lanes 1-8 respectively). The results show that MyTaq HS DNA Polymerase was more resistant to inhibition than that of supplier S. Individual colonies were picked from a plate of E.coli, washed directly into MyTaq buffer and amplified using MyTaq HS and primers for either a 2.6 kb or an 884 bp amplicon. Marker is HyperLadder 1kb (M). The results illustrate MyTaq DNA Polymerase is robust and gives highly–specific results.
|
Reagent |
200 Reactions |
1000 Reactions |
|
MyTaq HS Red Mix, 2x |
4 x 1.25 mL |
20 x 1.25 mL |
All components should be stored at -20°C upon receipt for optimum stability. Repeated freeze/thaw cycles should be avoided.
When stored under the recommended conditions and handled correctly, full activity of the reagents is retained until the expiry date indicated on the outer box label.
On Dry Ice or Blue Ice.
| Observation | Recommended Solution(s) |
| No or low PCR yield | Enzyme concentration too low – increase the amount of enzyme in 0.5 U increments. |
| Primers degraded – check quality and age of the primers. | |
| Magnesium concentration too low – increase concentration in 0.25 mM increments with a starting concentration of 1.75 mM. | |
| Primer concentration not optimized. Titrate primer concentration (0.3-1 µM); ensuring that both primers have the same concentration. | |
| Template concentration too low – Increase concentration of template. | |
| Perform a positive control to ensure that the enzyme, dNTPs and buffers are not degraded and/or contaminated. | |
| Multiple Bands | Primer annealing temperature too low. Increase annealing temperature. Primer annealing should be at least 5°C below the calculated Tm of primers. |
| Prepare master mixes on ice or use a heat-activated polymerase. | |
| For problems with low specificity. Try adding 3% DMSO (not supplied) to improve specificity. | |
| Smearing or artifacts | Template concentration too high. Prepare serial dilutions of template. |
| Too many cycles. Reduce the cycle number by 3-5 to remove non-specific bands. | |
| Enzyme concentration too high - decrease the amount of enzyme in 0.5 U increments. | |
| Extension time too long. Reduce extension time in 0.5-1 minute increments. |
Please click here in order to request your sample. You will receive an email confirmation within two business days with delivery details.