Ordering
*link will take you to our exclusive distribution partner site
*link will take you to our exclusive distribution partner site
ACCUZYME™ is a proprietary proofreading enzyme that offers increased-fidelity and high PCR yield, even in demanding applications. ACCUZYME has an error-rate of 3.0 x 10-6 and results in blunt-ended amplicons up to 5 kb in length, making it ideal for use in cloning and site-directed mutagenesis.
ACCUZYME is supplied with a buffering system that provides ideal conditions for most PCR assays. Consequently, the cost and effort typically associated with optimizing assay performance is often eliminated. In circumstances where further optimization is required to improve PCR specificity and/or yield, ACCUZYME includes an additional vial of MgCl2.
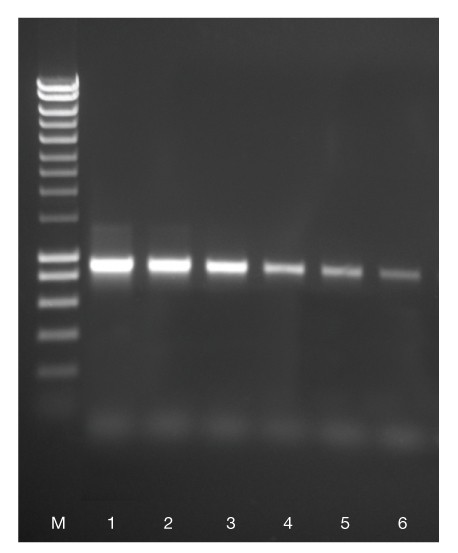
An 800 bp fragment was amplified from 500 ng, 50 ng, 5 ng, 0.5 ng, 50 pg and 5 pg (lanes 1-6 respectively) of human genomic DNA using ACCUZYME. HyperLadder 1kb (M). The result illustrates that ACCUZYME gives high product yield even from limiting amounts of template.
|
Reagent |
500 Reactions |
|
ACCUZYME DNA Polymerase |
200 µL |
|
10x AccuBuffer |
2 x 1.2 mL |
|
50 mM MgCl2 Solution |
1.2 mL |
2.5 u/µL
All components should be stored at -20°C upon receipt for optimum stability. Repeated freeze/thaw cycles should be avoided.
When stored under the recommended conditions and handled correctly, full activity of the reagents is retained until the expiry date indicated on the outer box label.
On Dry Ice or Blue Ice.
ACCUZYME DNA polymerase has an extremely fast extension rate compared to other proofreading enzymes, which are naturally slower than non-proofreading polymerases due to their ability to go back and correct nucleotide mis-incorporations. These polymerases will extend at a rate of as little as 30 s/kb, depending on the template amplified. Extension should be carried out at 68°C for optimal results.
| Observation | Recommended Solution(s) |
| No or low PCR yield | Enzyme concentration too low – increase the amount of enzyme in 0.5 U increments. |
| Primers degraded – check quality and age of the primers. | |
| Magnesium concentration too low – increase concentration in 0.25 mM increments with a starting concentration of 1.75 mM. | |
| Primer concentration not optimized. Titrate primer concentration (0.3-1 µM); ensuring that both primers have the same concentration. | |
| Template concentration too low – Increase concentration of template. | |
| Perform a positive control to ensure that the enzyme, dNTPs and buffers are not degraded and/or contaminated. | |
| Multiple Bands | Primer annealing temperature too low. Increase annealing temperature. Primer annealing should be at least 5°C below the calculated Tm of primers. |
| Prepare master mixes on ice or use a heat-activated polymerase. | |
| For problems with low specificity. Try adding 3% DMSO (not supplied) to improve specificity. | |
| Smearing or artifacts | Template concentration too high. Prepare serial dilutions of template. |
| Too many cycles. Reduce the cycle number by 3-5 to remove non-specific bands. | |
| Enzyme concentration too high - decrease the amount of enzyme in 0.5 U increments. | |
| Extension time too long. Reduce extension time in 0.5-1 minute increments. |
Please click here in order to request your sample. You will receive an email confirmation within two business days with delivery details.