Ordering
*link will take you to our exclusive distribution partner site
*link will take you to our exclusive distribution partner site
The ISOLATE II RNA Mini Kit provides a simple, efficient column-based method for the isolation of total RNA from a wide variety of starting materials, without the need for hazardous reagents such as phenol.
By combining the stringency of guanidinium-thiocyanate lysis with the speed and ease-of-use of silica-membrane purification, the ISOLATE II RNA Mini Kit provides a fast method for the purification of high-quality total RNA from animal and plant cells and tissues as well as cultured cells, bacterial cells, yeast, biological fluids and cell-free samples.
Biological samples which are sometimes difficult to process i.e. mouse tissue (liver, brain), various tumor cell lines, Streptococcus and Actinobacillus pleuropneumoniae, will yield high-quality RNA with the ISOLATE II RNA Mini Kit.
The online product manual has protocols for purifying total RNA from cultured cells, tissues, yeast, bacteria, biological liquids, paraffin embedded tissue and RNAlater® treated samples. There is also a protocol for a convenient on-column DNase treatment, using RNase-free DNase I that is supplied with the kit, for applications that are sensitive to very small amounts of DNA.
The ISOLATE II RNA Mini Kit has been designed to deliver optimal performance in RT-qPCR in conjunction with either the SensiFAST cDNA Synthesis Kit and SensiFAST Real-Time PCR Kits, or the SensiFAST One-Step Real-Time RT-PCR Kits. Additionally, the ISOLATE II RNA Mini Kit can be used to purify samples for PCR and RT-PCR amplification using the Tetro cDNA Synthesis Kit and any enzyme from the Meridian PCR portfolio, including MyTaq DNA Polymerase.
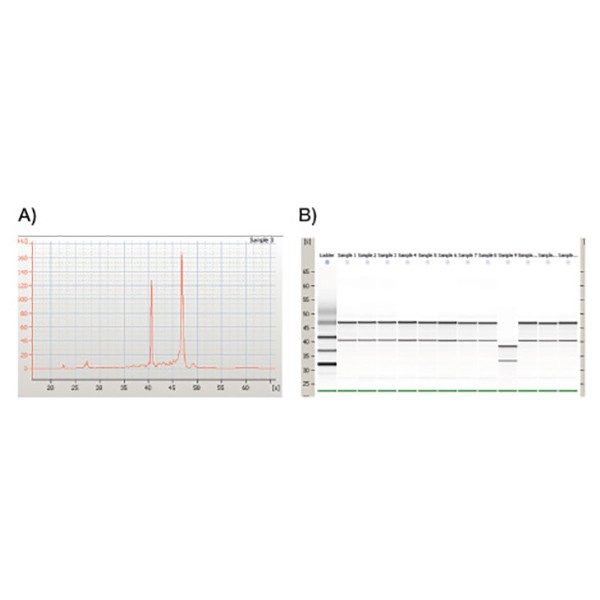
High quality RNA
RNA was isolated from HeLa cells using ISOLATE II RNA Mini Kit and analyzed by the Bioanalyzer 2100 (Agilent Technologies). The results illustrate A) The quality of RNA, which was found to be exceptional (RIN: ≥9.2) and B) highly reproducible across samples.
Superior performance in real-time applications
RNA was isolated in a 10-fold serial dilution (14,000, 1,400, 140, 14 and 1.4 cells), from mouse 3T3 cells (lanes 1-5 respectively), using ISOLATE II RNA Mini Kit (red traces) and an equivalent kit from Supplier Q (green traces). Subsequently, real-time reverse transcriptase reactions were performed using SensiFAST SYBR No-ROX One-Step Kit. The results illustrate the higher quality of the extraction process over several orders of magnitude, down to a few cells.
|
Reagent |
10 Preps |
50 Preps |
250 Preps |
|
ISOLATE II Filters |
10 |
50 |
250 |
|
ISOLATE II RNA Mini Columns & Collection Tubes |
10 |
50 |
250 |
|
Collection Tubes (2 mL) |
30 |
150 |
750 |
|
Collection Tubes (1.5 mL) |
10 |
50 |
250 |
|
Lysis Buffer RLY |
10 mL |
25 mL |
125 mL |
|
Wash Buffer RW1 |
15 mL |
15 mL |
80 mL |
|
Wash Buffer RW2 |
6 mL |
12 mL |
3 x 25 mL |
|
Membrane Desalting Buffer MEM |
10 mL |
25 mL |
125 mL |
|
Reaction Buffer for DNase I RDN |
7 mL |
7 mL |
30 mL |
|
DNase, RNase-free (lyophilized) |
1 Vial |
1 Vial |
An increase of A230 values may be caused by different substances, like carbohydrates, peptides and phenol. A bad A230/A260 ratio in RNA samples is mostly due to a contamination with guanidinium thiocyanate which is present in several reagents used for RNA extraction, for instance in the lysis buffer. In contrast to a bad A280/A260 ratio it does not automatically reflect a bad RNA quality. Currently there is no consensus about a lower limit of this ratio and mostly a carry-over of guanidinium thiocyanate does not affect the reliability of downstream applications. Nevertheless, an extra washing step with RW2 would be helpful to avoid this problem. And it would be helpful to pipet the flow-through out of the collection tube, instead of pouring it off.
For the optimal recovery of large RNA/DNA fragments it is necessary to optimize the elution step. To increase the recovery rate it would be helpful to use a larger volume for elution, incubate the column with the elution buffer prior to centrifugation and use repeated elution steps. To avoid excessive dilution it may be helpful to reapply the eluted fraction again.
Agarose gel analysis of RNA samples can be both valuable and misleading. The pattern of bands on the gel can not only be indicative of the quality of the sample, but equally it can also only indicate that the gel tank, buffer or agarose is contaminated with RNase. A safer measure is to use a Bioanalyzer and look at the RIN value (RNA Integrity Number), which should be as close to 10 as possible, indicating that the RNA is not degraded. A spectrophotometer (ideally a microfluidic one) can also be used to determine the ratio of A260 to A280 for purity determination.
All these methods give an idea of the quality of total RNA which tends to be dominated with rRNA and tRNA. It is not uncommon for an agarose gel to show an abundance of 18S and 23S rRNA but on analysis for there to be little of the transcript of interest. Ideally RNA quality control should include an assessment of the presence of common transcripts (from reference genes, such as GAPDH) using RT-PCR or RT-qPCR.
If the yield of RNA is low, it is best to first check that all the solutions used and the equipment employed is largely free of RNase. Solutions should be prepared with the highest quality reagents available, preferably ones that have a certificate of analysis to show that they are free of DNase and RNase. If there is confidence that RNase contamination is at a minimum, then it may be worth ensuring that the sample is properly homogenised before lysis and to check a sample of the lysed material under a light microscope to ensure that all the cells are disrupted.
RNA samples that have been contaminated with RNase (either during processing or those that have been sent from another lab) can be purified using the ISOLATE II RNA Micro Kit.