Ordering
*link will take you to our exclusive distribution partner site
*link will take you to our exclusive distribution partner site
Formerly RNA Extraction Control 670.
RT-qPCR Extraction Control not only enables users of a diagnostic qPCR assay to determine if there are inhibitors in the PCR assay, but also to validate the success of the extraction step, reducing the chance of obtaining a false negative result in the sample RNA.A common practice in qPCR is to add a known amount of spiked control RNA after RNA extraction, this monitors PCR inhibition but has no value as an extraction control. The ideal situation is to have the test sample and internal control undergo the same processing prior to qPCR. Meridian has developed a RT-qPCR Extraction Control, which more closely mimics the test sample, as compared to spike controls. Genetic material from the test sample and the RT-qPCR Extraction Control is simultaneously extracted by common extraction methods, with the extraction control being as sensitive to inhibition and extraction failure as the test sample.
Artificial RT-qPCR Extraction Control cells are of a known concentration, containing the Internal Control RNA sequence. This sequence contains no known homology to any organism and, importantly, has minimal interference with detection of sample RNA. The RT-qPCR Extraction Control cells are spiked into the lysis buffer with the target sample, prior to RNA extraction. Control Mix, which includes primers and probe, is then added to the reaction mix before amplification. Signal derived from the Internal Control RNA confirms the success of the extraction step. RT-qPCR Extraction Control also monitors co-purification of PCR inhibitors that may cause biased or false amplification patterns.
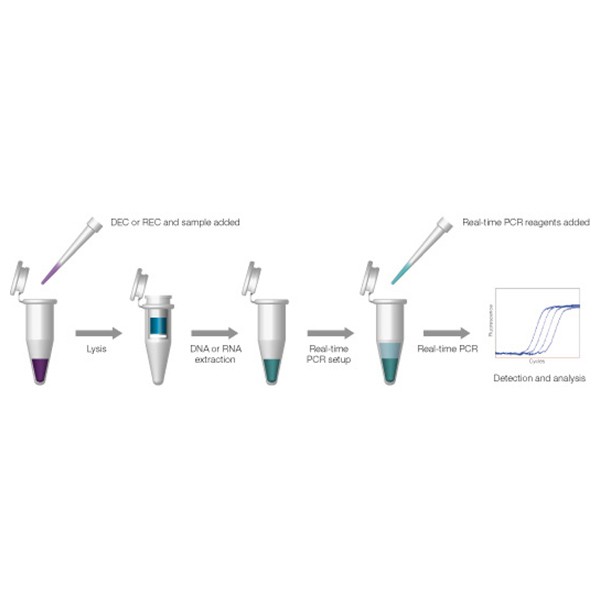
Illustration of the extraction process
REC assesses effects of extraction as well as inhibition throughout the entire workflow.
REC monitors inefficient RNA extraction
ISOLATE II RNA Mini kit was used to extract the RNA from HeLa cells containing A) spiked control DNA and B/ REC. To show inefficient extraction, the lysis buffer and/or binding buffer substituted with PBS. The results demonstrate complete lysis (yellow line), no lysis buffer (brown), no binding buffer (grey), no lysis and no binding buffer (pink). The results illustrate that the spiked control DNA is insensitive to extraction failure, whereas REC is sensitive, with the Ct values being affected.
PCR reaction inhibition
ISOLATE II RNA Mini kit was used to extract the RNA from HeLa cells containing REC. Increasing concentrations of EDTA were included in the reaction to simulate increasing levels of inhibition. The results illustrate that REC is increasingly inhibited by increasing concentrations of EDTA, showing that inhibition of PCR reaction can be identified using REC.
RT-qPCR Extraction Control Red is suitable for use with commercially available silica-membrane RNA extraction kits and CHELEX matrices and has been tested on a wide range of qPCR platforms including ABI-7500, LightCycler 480®, RotorGene-Q™, Mic and MX3005P®.
RT-qPCR Extraction Control Red uses Quasar® 670 and is also available with Cal Fluor® Orange 560 to fit in with existing protocols. CAL Fluor and Quasar dyes are performance-optimized fluorophores for multiplex qPCR.
|
Reagent |
500 Reactions |
|
Internal Control RNA Red |
5 x 200 µL |
|
25 x Control Mix 670 |
5 x 100 µL |
All kit components should be stored at -80°C upon receipt. When stored under the recommended conditions and handled correctly, full activity of the kit is retained until the expiry date on the outer box label.
Shipped on dry ice.