Ordering
*link will take you to our exclusive distribution partner site
*link will take you to our exclusive distribution partner site
MyTaq™ One-Step RT-PCR Kit incorporates the latest advances in buffer chemistry, including Meridian ultra-pure dNTPs, together with a proprietary reverse transcriptase and MyTaq HS, a new generation of antibody-mediated hot-start DNA polymerase. This ensures that MyTaq One-Step RT-PCR Kit enables fast, highly-specific and ultra-sensitive amplification of RNA targets for use in a broad range of downstream applications.
MyTaq One-Step Kit is ideal for determining the presence or absence of RNA templates and quantifying expression through qualitative or semi-quantitative analysis of RNA transcription levels. The one-step format uses gene specific primers to maximize amplification of these targets, whilst eliminating non-specific amplification, ensuring highly efficient and sensitive transcription from as little as 3 pg total RNA.
MyTaq One-Step RT-PCR Kit is perfect for the synthesis of double-stranded cDNA products for subsequent gene expression analysis over a broad temperature range, the use of higher temperatures allows reverse transcription through RNA secondary structure, including difficult and GC-rich sequences. Enhanced amplification of these targets ensures high cDNA yields from all RNA, including total RNA, mRNA, in vitro transcribed RNA, snRNA and viral RNA.
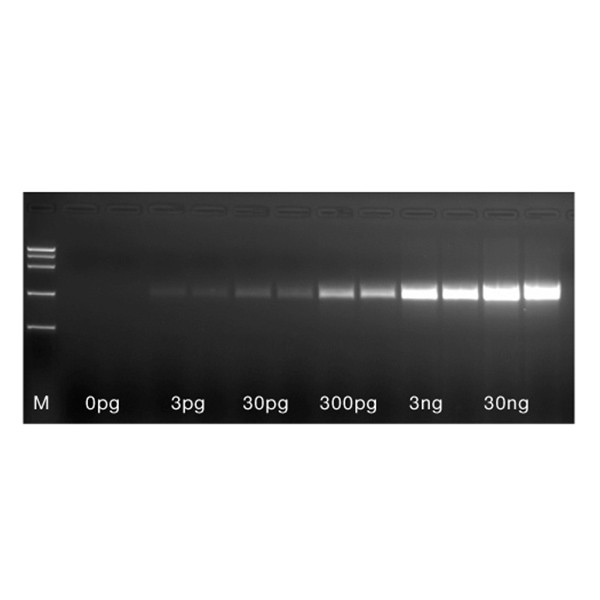
A 1 kb fragment was amplified in duplicate from serial dilution of mouse total RNA (30 ng, 3 ng, 300 pg, 30pg and 3 pg respectively) using RN18S-1000 primers and the MyTaq One-Step RT-PCR Kit. The completed RT-PCR reactions were analysed on a 1% agarose gel. HyperLadder 50bp (M). The results show that the MyTaq One-Step RT-PCR Kit achieves high–yield, specific amplification from even complex templates.
A 1 kb fragment was amplified in duplicate from a serial dilution of mouse total RNA (10 ng, 2 ng, 400 pg, 80 pg, 16 pg and 3 pg; lanes 1-6 respectively) using RN18S-1000 primers and the MyTaq One-Step RT-PCR Kit. HyperLadder 50bp (M). The reverse transcriptase in the MyTaq One-Step RT-PCR Kit was able to deliver high quality cDNA even at 50°C, over a broad dynamic range.
|
Reagent |
25 Reactions |
100 Reactions |
|
MyTaq One-Step mix (2x) |
1 x 625 µL |
2 x 1.25 mL |
|
RiboSafe RNase Inhibitor (10 u/µL) |
1 x 25 µL |
1 x 100 µL |
|
Reverse transcriptase |
1 x 12.5 µL |
1 x 50 µL |
|
DEPC-treated Water |
1 x 1.8 mL |
1 x 1.8 mL |
All components should be stored at -20°C upon receipt for optimum stability. Repeated freeze/thaw cycles should be avoided.
When stored under the recommended conditions and handled correctly, full activity of the reagents is retained until the expiry date indicated on the outer box label.
Shipped on dry/blue ice
| Observation | Recommended Solution(s) |
| No or low PCR yield | Enzyme concentration too low – increase the amount of enzyme in 0.5 U increments. |
| Primers degraded – check quality and age of the primers. | |
| Magnesium concentration too low – increase concentration in 0.25 mM increments with a starting concentration of 1.75 mM. | |
| Primer concentration not optimized. Titrate primer concentration (0.3-1 µM); ensuring that both primers have the same concentration. | |
| Template concentration too low – Increase concentration of template. | |
| Perform a positive control to ensure that the enzyme, dNTPs and buffers are not degraded and/or contaminated. | |
| Multiple Bands | Primer annealing temperature too low. Increase annealing temperature. Primer annealing should be at least 5°C below the calculated Tm of primers. |
| Prepare master mixes on ice or use a heat-activated polymerase. | |
| For problems with low specificity. Try adding 3% DMSO (not supplied) to improve specificity. | |
| Smearing or artifacts | Template concentration too high. Prepare serial dilutions of template. |
| Too many cycles. Reduce the cycle number by 3-5 to remove non-specific bands. | |
| Enzyme concentration too high - decrease the amount of enzyme in 0.5 U increments. | |
| Extension time too long. Reduce extension time in 0.5-1 minute increments. |